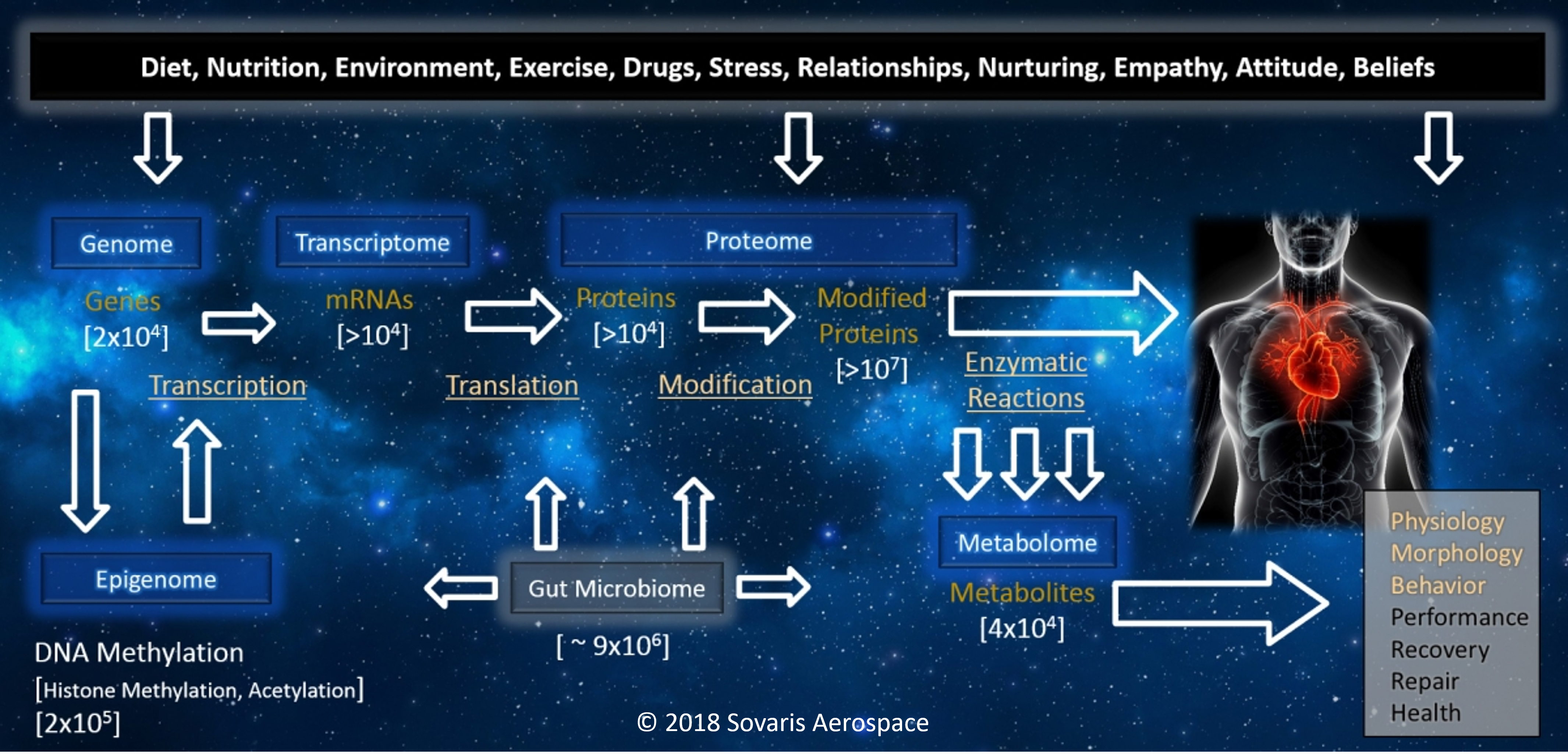
The Omics Landscape
Our work incorporates a combination of targeted and untargeted omics, which can be used to better understand the dynamics involved in the conditions we study. Each of the primary Omics disciplines below has the capacity to reveal unique patterns of molecular variance under performance, health, and disease conditions, which can inform countermeasure parameters, and aid in their development.
Genomics
Assess the DNA exome, including single nucleotide polymorphisms, copy number variants, insertions, and deletions. SNPs can inform investigators about susceptibility to various conditions and aid in determination of whether there is an optimum or limiting genotype (single genes or clusters of genes) for a given condition.
Epigenomics
Transcriptomic regulating factors not empirically coded in the genomic sequence. Epigenetic patterns under various conditions may aid in identification of gene regulating effects that impact the body’s response.
Transcriptomics
Assessment of gene expression through RNA transcripts. Gene expression patterns under various conditions may aid in detecting different patterns of change over time. This may yield new potential assessment targets and lead to new countermeasure targets.
Proteomics
Assessment of protein synthesis and post-translational protein modification. Protein expression patterns under various conditions may aid in optimizing countermeasure parameters.

Metabolomics
Assessment of the small molecule pool (below 1500 amu). Metabolite patterns may reveal changes across metabolic networks under varied conditions. Metabolomics is used in targeted and non-targeted ways for detection and hypothesis generation, respectively.

Exposomics
Assessment of the exposome. The exposome is the sum of environmental exposures and the response to those exposures. The full complement of integrated omics can be used to describe the human response to these exposures.

Metallomics
Assessment of the entirety of metals and metalloids within a cell or tissue type. Broadly speaking, metallomics is the study of the metallome, interactions, and functional connections of metal ions and other metal species with genes, proteins, metabolites, and other biomolecules in biological systems.

Integrated Omics
Targeted integrated omics is used to assess molecular profiles of individuals, under our personalized medicine efforts. This approach to targeted integrated omics commonly includes, 1) genomics (selected SNPs), 2) metabolomics (dozens to hundreds of small molecules), and 3) proteomics (selected proteins specific to the mission, performance, or health objective).
Microbiome
Assessment of the gut microbiome composition at the phylum and species level, using genomics. This microbial genome is estimated to contain 4-9 million genes and produce a complex array of small molecules, some of which are absorbed intact and influence human metabolism. Gut genome and metabolome (targeted and untargeted) data are correlated to better understand the physiological response, and to explore new solutions that affect health and performance.
Continuum of Omics for Spaceflight Medicine, Clinical Medicine, and Research on Earth and in Space